About me
I am a Machine Learning Engineer at Affirm since 2024 August.
I am also a 7th-year Ph.D. student in Jian Ma's lab in the School of Computer Science at Carnegie Mellon University. I passed my thesis defense on June 5, 2024. I earned a Master in Machine Learning during my Ph.D. and hold a B.E. in Computer Science and Technology from Yao Class, Institute for Interdisciplinary Information Sciences at Tsinghua University. During undergraduate, I worked with Prof. Jianyang Zeng at Tsinghua University and Prof. Jinbo Xu at Toyota Technological Institute at Chicago.
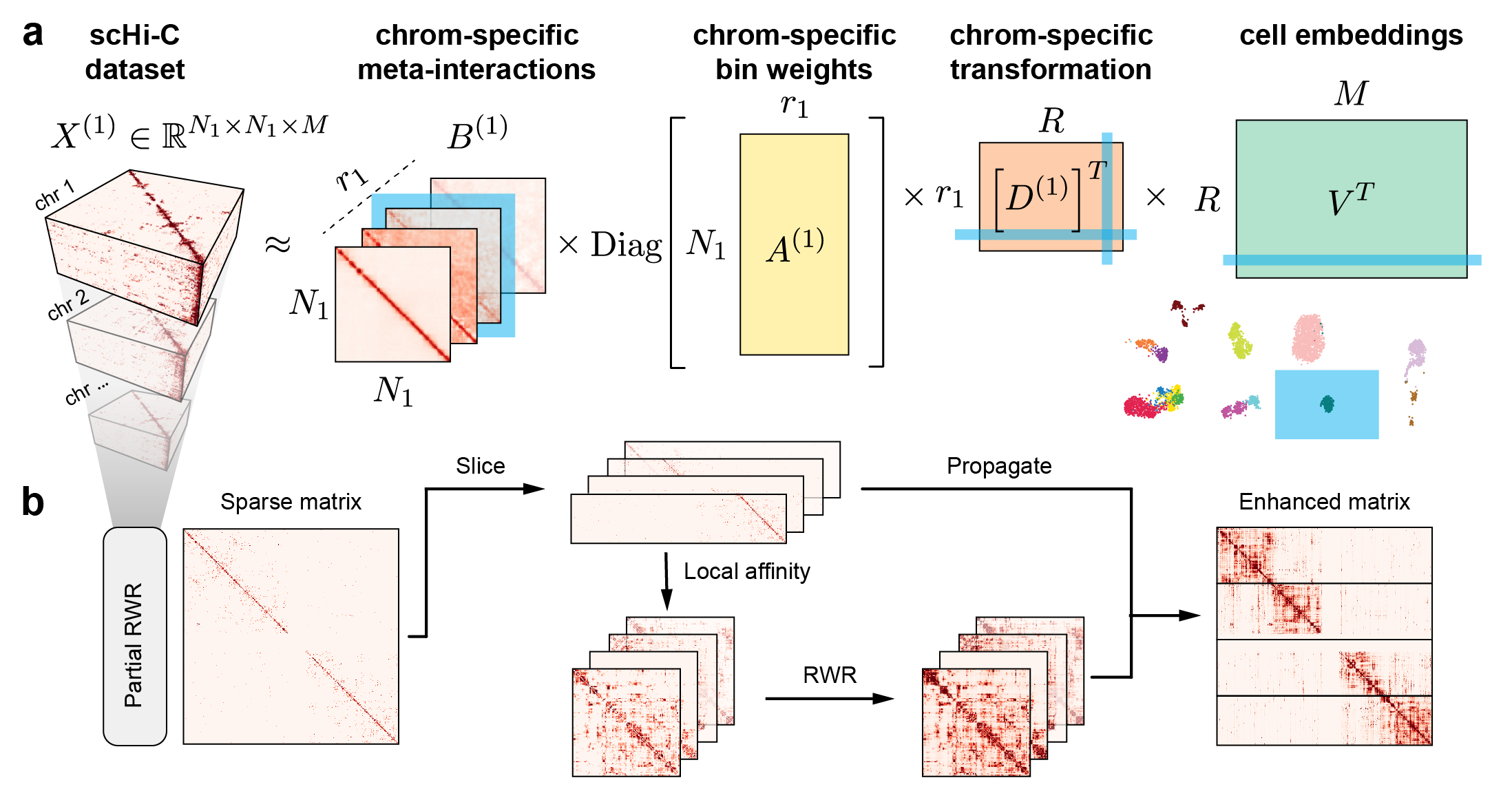
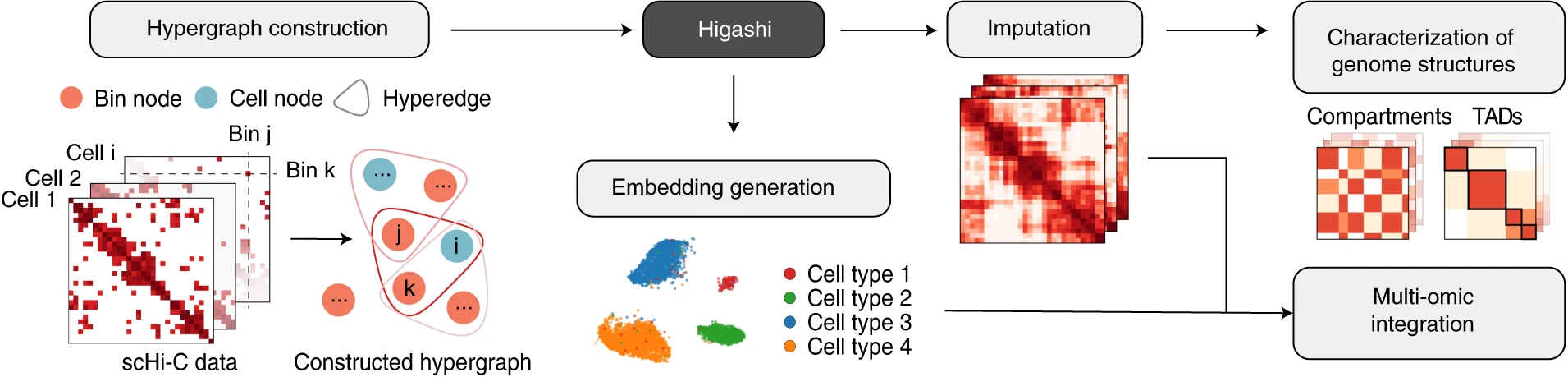
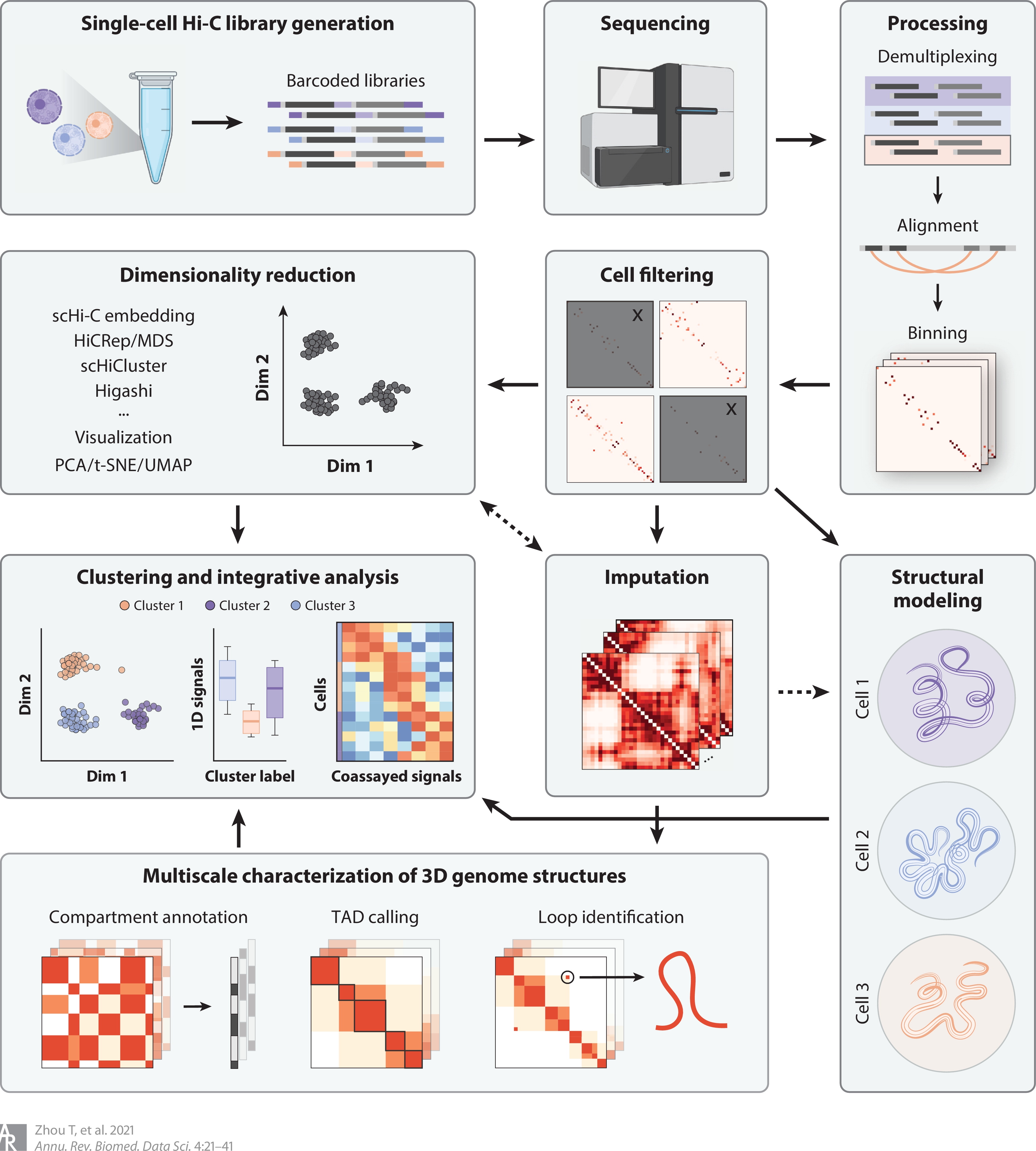
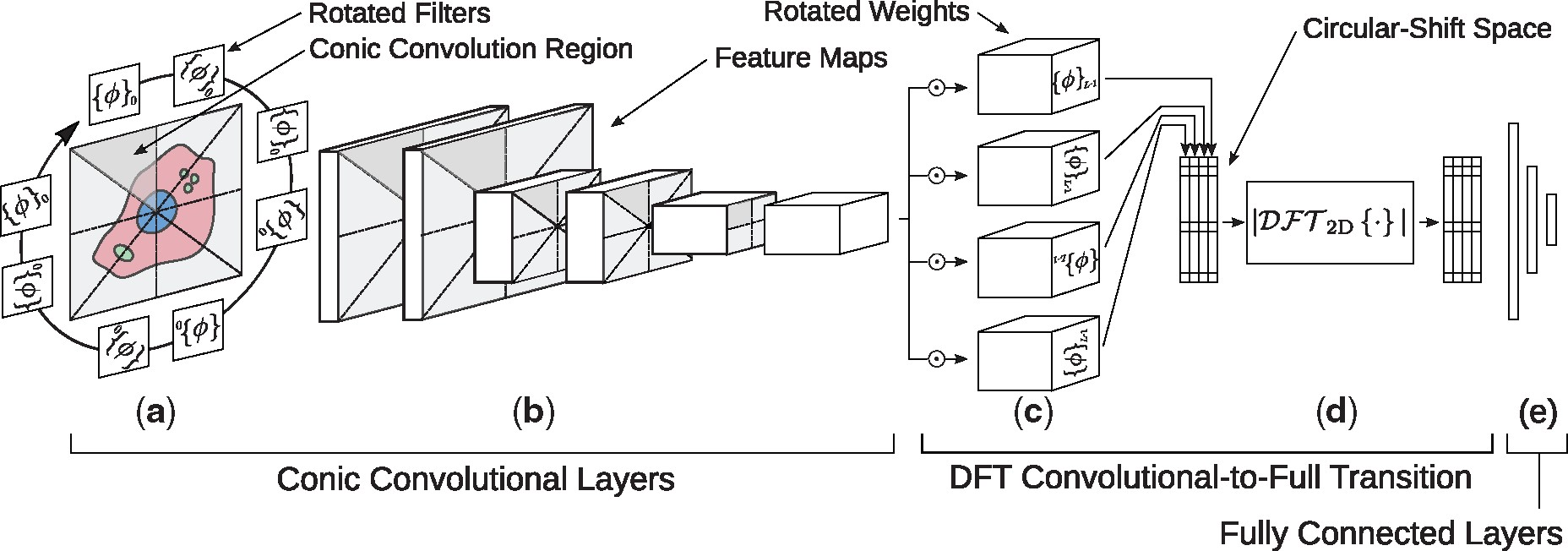
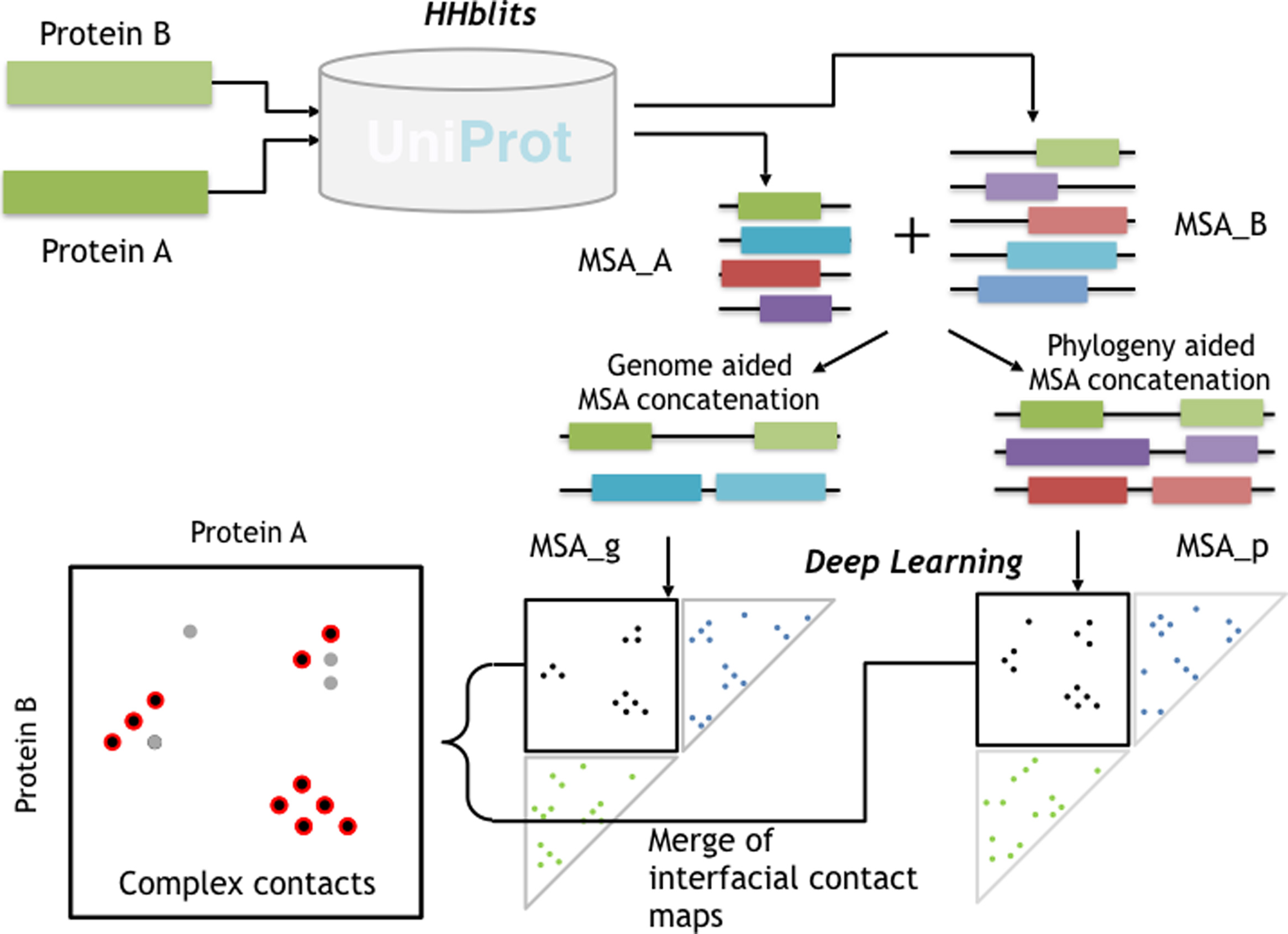
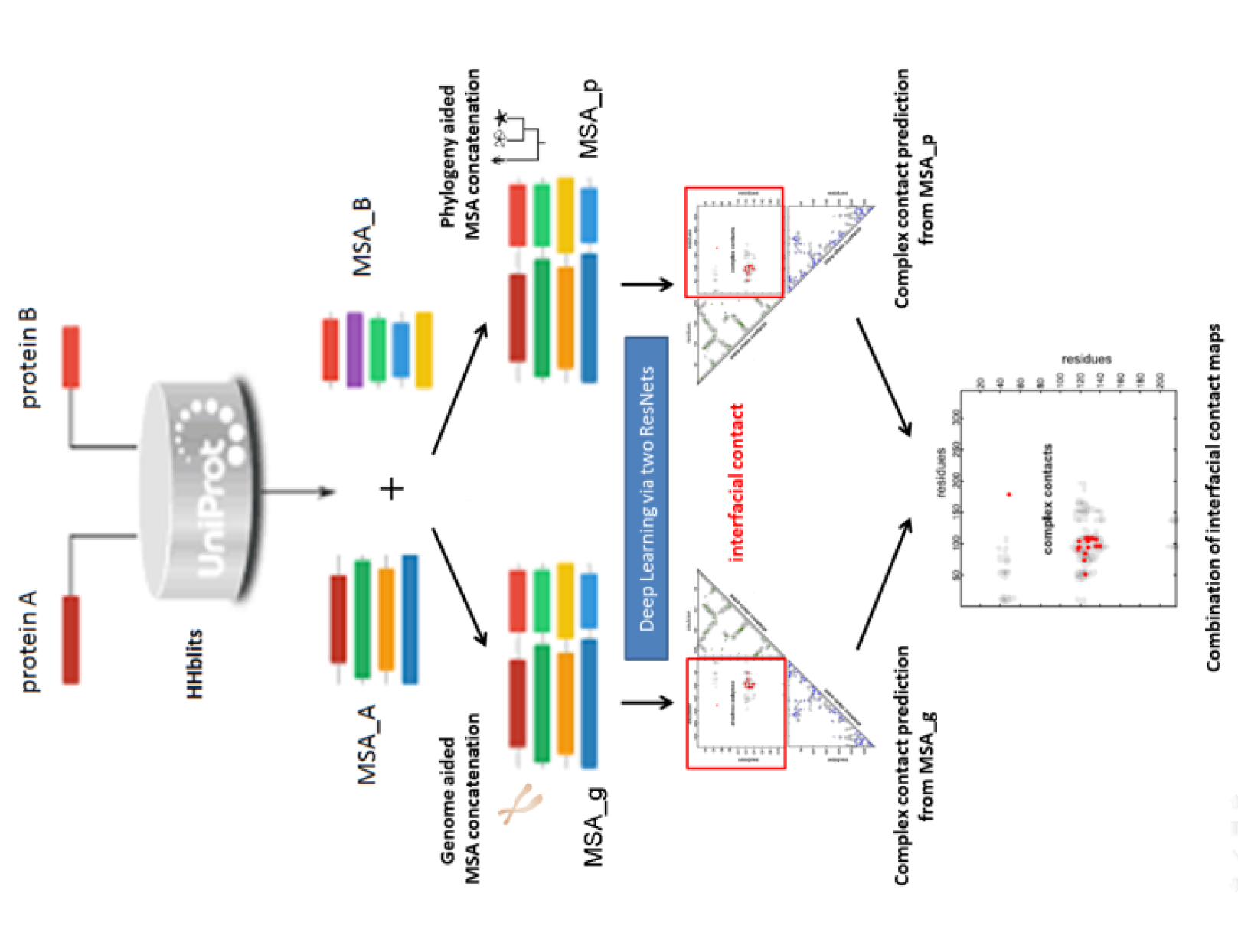
My research passion resides at the intersection of multimodal representation learning and biology. I am dedicated to advancing the field through the development of explainable artificial intelligence (XAI) methodologies, including probabilistic graphical models, graph neural networks, large language models, and tensor decomposition algorithms. Furthermore, I specialize in interpreting model outcomes and exploring cross-modality relationships. I apply these methodologies to tackle real-world challenges in various biological areas, including spatial transcriptomics, 3D genome structures, single-cell multiome, and protein-protein interactions.